Bioinformatics against viruses on the example of SARS-CoV-2
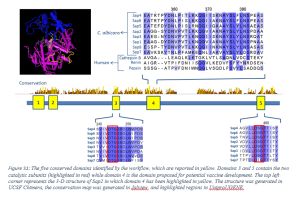
At the end of July 2020, group of American scientists published the article on the development of new computational bioinformatics workflow that helps to identify therapeutic or vaccine targets against dangerous viruses. They demonstrated the example on the coronavirus SARS CoV-2, which causes the disease COVID-19, explaining all the steps of the workflow. The authors describe how the workflow can be adapted to other infectious agents. It is important that the workflow is accessible to a wide range of students regardless of their initial bioinformatics skills, and is based on freely available software. We are delighted to know that our open platform UGENE was chosen for this new computational workflow and now is used in two of its six phases, along with three other bioinformatics systems. UGENE helps to visualize and analyze the alignments of viral genomic sequences and to filter candidate sequences. Along with delivering the specific results, new workflow helps students to reinforce several concepts of medical virology.
UGENE has good history of successful usage by virology scientists in different parts of the world, helping in the identification of new viruses, their comparative analysis and search for new methods to tackle dangerous infections.

0 Comments